
But even after I obtain it, I still need to use more tricks (such as blending, or bending) in order to tune-up my G matrix to be invertible and well-conditioned. First, I have to deal with many tricks on pre-processing my molecular data (often SNPs) in order to calculate my G matrix. My first surprise was the level of complexity associated with the generation of the G matrix. So, the process seems simple and straightforward, but it is not! The theory is simple: just replace your numerator relationship matrix A with a genomic-based relationship matrix G and you are now doing what is known as GBLUP. Now with the genomics era here, I have been moving from this pedigree-based animal model to a genomic-based animal model. Nowadays, this takes even less time, given that computers have more memory and computing power.
This, back then, took a few minutes, and I was always excited to see a large file with all the breeding value estimates.

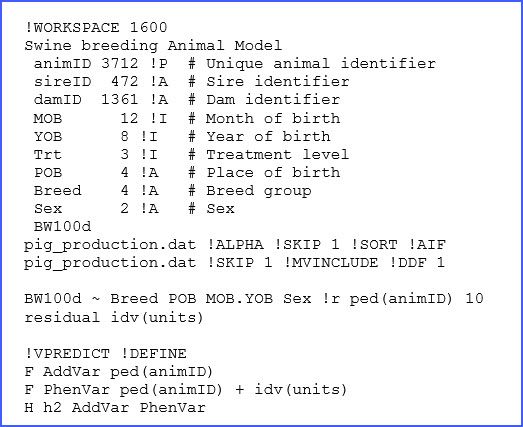
I started in the pre-genomics era and I recall fitting an animal model during my PhD with more than half a million records in my pedigree file without any issues. I have been a user of ASReml-R for more than 10 years.


 0 kommentar(er)
0 kommentar(er)
